|
Singapore-MIT Alliance for Research & Technology |
|||||||||||||||||||||
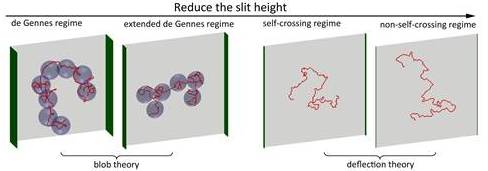
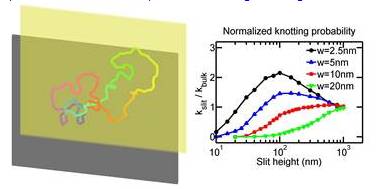
Research Projects Under Thrust 1: Molecular Scale ToolsPatrick DOYLE (MIT), Matthew LANG (MIT), Bruce TIDOR (MIT), Krystyn VAN VLIET (MIT), Johan van der Maarel (NUS), Lars Nordenskiöld (NTU), Zhisong WANG (NUS), Qing Hua XU (NUS), Jie YAN (NUS) In Thrust 1, BioSyM’s molecular manipulation tools include a suite of capabilities that are focused on high-resolution measurement of how and why biomacromolecules interact under crowded, confined, and mechanically strained conditions common in biological systems. Our force probe arsenal includes custom built magnetic, optical, and atomic force microscopes, many of which have been integrated within high-end multi-dimensional imaging platforms capable of simultaneous measurement of force, color, and time. Specific projects include nanofluidic studies of DNA structure and stability; rheology of biogels as related to health and disease; novel approaches to drug target identification through computer-modeling enabled analysis of biological pathways; biological-partbased synthetic materials, structures, sensors, and machinery. These studies will advance our understanding of the foundational principles of molecular interactions and bio-systems integration1.Conformation and Dynamics of DNA in Nanofluidic DevicesDai Liang (SMART), Jeremy Jones (MIT), Benjamin Renner (MIT), Patrick Doyle (MIT/SMART), Jie Yan (NUS) and Johan van der Maarel (NUS) This project is to study the conformation and dynamics of DNA in nano-confinement, e.g. nano-channels and nano-slits. The understanding of DNA behaviour will benefit the development of nanofluidic devices for genome mapping. The major methodology is Monte-Carlo simulation of DNA in confinement. First, we systematically study DNA conformation in slit-like confinement from weak to strong confinement. The simulation results agree with experiments, and provide new insights to the transition regime from weak to strong confinement. Secondly, we study the knotting probability of circular DNA in confinement. We find that the knotting probability may vary with the slit height monotonically or non-monotonically, depending on the contour length, effective chain width (ionic strength dependent) and persistence length of DNA. This result will guide future experiments which target to produce populations of knotted DNA. Thirdly, DNA conformations in square channels (or tubes) are successfully described by blob theory and deflection theory in moderate and strong confinements, respectively. We develop a new theory to bridge the gap between weak and strong confinements, and validate the theory by simulation results. Finally, we are studying the diffusivity of DNA in slitlike confinement. The motivation is to discover the reason why the scaling of diffusivity is different between experiments and blob theory, as well as to understand the diffusivity in the transition regime. Overall, our research deepens the understanding of DNA behaviours in nano-confinement, especially in the transition regime (channel width ~ 100 nm).
Selected publications
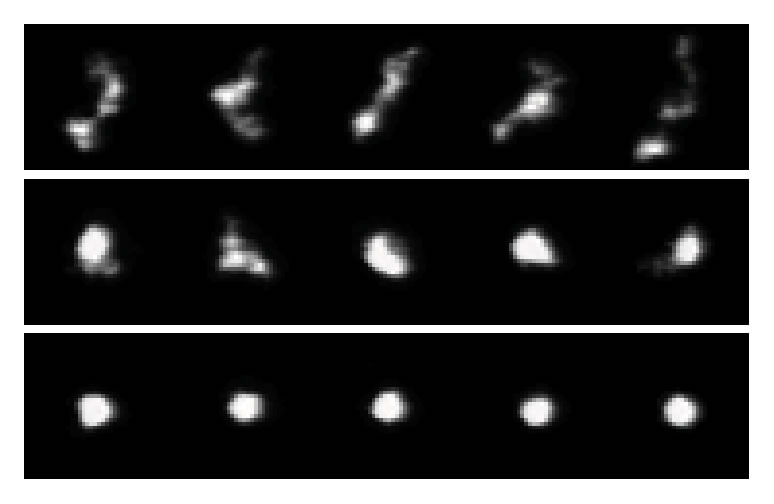
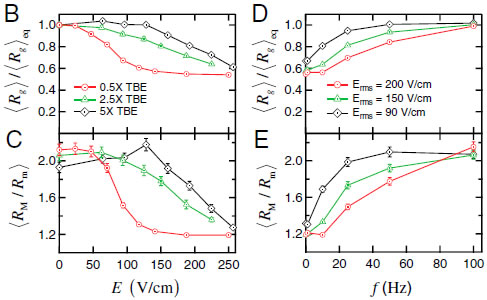
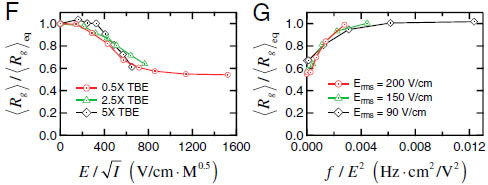
2.Compression and self-entanglement of single DNA molecules under electric fieldNing Du (SMART), Jing Tang (MIT), Jeremy J. Jones (MIT), Ben Renner (MIT), Patrick Doyle (MIT/SMART), Jie Yan (NUS) and Johan van der Maarel (NUS) We experimentally study the effects of a uniform electric field on the conformation of single DNA molecules. We demonstrate that a moderate electric field (∼200 V∕cm) strongly compresses isolated DNA polymer coils into isotropic globules. Insight into the nature of these compressed states is gained by following the expansion of the molecules back to equilibrium after halting the electric field. We observe two distinct types of expansion modes: a continuous molecular expansion analogous to a compressed spring expanding, and a much slower expansion characterized by two long-lived metastable states. Fluorescence microscopy and stretching experiments reveal that the metastable states are the result of intramolecular self-entanglements induced by the electric field. These results have broad importance in DNA separations and single molecule genomics, polymer rheology, and DNA-based nanofabrication.
Selected publications
3. The structures and functions of DNA under tension Xinghua Zhang (SMART), Peiwen Cong (NUS), Patrick Doyle (MIT/SMART), Jie Yan (NUS) and Johan van der Maarel (NUS)
Double-stranded DNA is a dynamic molecule whose structure can change depending on conditions. While there is consensus in the
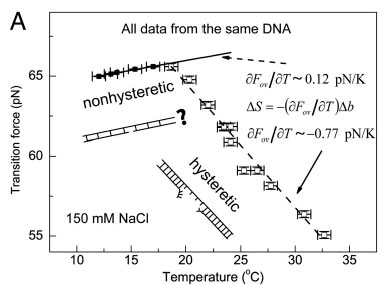
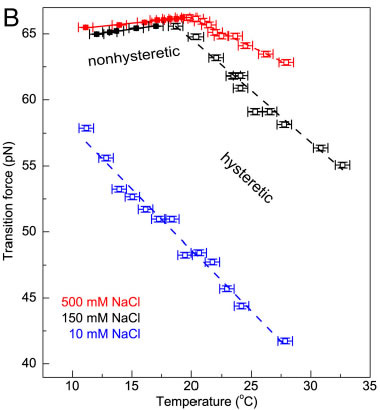
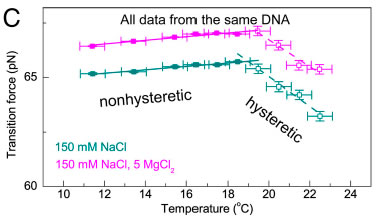
Measurements of Fov(T) including error bars in both force and temperature. In the nonhysteretic transition, Fov is denoted by colored filled squares and fitted to a linear function (solid line). In the hysteretic transition, Fov is denoted in colored open squares, and it is fitted to a linear function (dashed line). (A) Fov(T) in 150 mM NaCl and pH 7.5 (black). A linear relation with a slope of approximately 0.12 pN∕K was determined in the nonhysteretic transition (T < 18 °C). The slope remains unchanged if the midpoint or the terminus of the transition was used to determine the transition force (SI Appendix, Determination of the transition force). A different linear relation with a slope of −0.77 pN∕K was determined in the hysteretic transition (T > 18 °C). The hysteretic transition has been agreed to be peeling of one strand from the other, while the nonhysteretic transition leads to an undetermined DNA structure, which are illustrated in the panel. (B) In 500 mM NaCl (red), a similar piecewise linear relation was obtained with a slope of approximately 0.10 pN∕K in the nonhysteretic transition and approximately −0.44 pN∕K in the hysteretic transition. In 10 mM NaCl (blue), only the hysteretic transition occurred and a single linear region was observed with a slope of approximately −0.92 pN∕K. The data obtained in 150 mM (A) are also plotted for comparison (black). The effects of Mg2+ to DNA overstretching in 150 mM NaCl and pH 7.5.Without Mg2+ (dark cyan), switching from the nonhysteretic transition with a slope of approximately 0.08 pN∕K to the hysteretic transition with a slope of approximately −0.68 pN∕K occurred at approximately 18.5 °C. In the presence of 5mM Mg2+ (magenta), switching fromthe nonhysteretic transition with a slope of approximately 0.08 pN∕K to the hysteretic transition with a slope of approximately −0.62 pN∕K occurred at approximately 19.4 °C. Selected publications
4. Multispectral Force-Fluorescence Microscope Siew-Kit Hoi (SMART), Taishi Zhang (NUS), Zhenping Guan (NUS), Matt Lang (MIT/SMART), Patrick Doyle (MIT/SMART), Yan Jie (NUS) and Qing-Hua Xu (NUS)
We have developed an integrated fluorescence and force (through: optical, fluidic, magnetic) microscope appropriate for single molecule and cell studies. Custom machining and general assembly of microscope were completed at MIT, and the entire system transferred and commissioned at BioSyM SMART' opto-mechanical suite in CREATE. The instrumentaion features double trap geometry. The combination of the optical tweezers microscopy and single molecule fluorescence spectroscopy has provided the capability for simultaneous manipulation and monitoring of molecular structure in real time. The optical traps can be employed to supply controlled external loads while fluorescent molecules report the concurrent information about macromolecular structure.
Selected publications
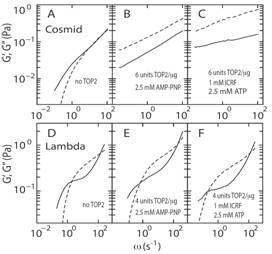
5. Gelation of genome by topoisomerase II targeting anticancer agentsYun Soo Kim (SMART), Binu Kundukad (SMART) Abdollah Allahverdi (NTU), Lars Nordenskold (NTU), Patrick S. Doyle (MIT/SMART) and Johan R. C. van der Maarel (NUS) Topoisomerase II (TOP2) regulates the topology of DNA by catalysis of a double strand passage reaction. Inhibition of this reaction prevents cell replication, and, thus, is a pathway targeted by anticancer drugs. Some details regarding the cell-killing mechanism are unknown and assays to screen for anticancer drugs are not well established. Here, we study the gelation of linear and circular DNA using microrheology assays. Gelation of the DNA–enzyme mixture was examined by tracking of multiple colloidal probe particles. The mean square displacements of the probe particles were analyzed by the time-cure superposition procedure as well as the classical derivation of the dynamic moduli. First, the passage reaction was inhibited by AMP-PNP, a non-hydrolyzable analog of ATP. The results showed gelation due to the formation of a self-catenated network of circular DNA molecules. Next, when TOP2 was inhibited by the anti-cancer drug ICRF-193, we observed a similar change in rheology. Based on these findings, we propose a cell-killing mechanism by gelation of the genome through TOP2-mediated interlocking of looped DNA segments of the replicated, intertwined chromosomes.
Selected publications
6. Understanding Molecular Basis of Mechanotransduction using Computer Simulations(Bruce Tidor (MIT/SMART-BioSyM) and Sudipta Samanta (SMART-BioSyM)At the molecular level, force induced conformational change can alter protein activity; one possible mechanism for altering activity is through changes in chemical properties such as binding affinity and enzyme activity. In this framework, force-induced alteration of binding affinity, as but one example, would depend on characteristics of the applied force, including direction, magnitude, and point of application. Currently the property space of how applied force affects chemical properties like binding affinity is poorly understood due to the lack of effective experimental data. Here we develop a computational approach and analysis for gaining insight into force modulated binding properties. The main goal in this project is to understand the molecular basis of mechanotransduction. For this we apply free energy simulation to study the binding response to applied force of focal adhesion targeting domain of the focal adhesion kinase bound to the paxillin motif. Applied force on the protein in different directions helps understand how its direction and magnitude can affect structure-function relationship in biologically relevant systems. 7. Robust inhibition of Hepatitis C viral propagation(Bruce Tidor (MIT/SMART-BioSyM), Pradeep Anand Ravindranath (NUS) and Nathaniel W.Silver (MIT)The goal is to design mutation resistant inhibitors for hepatitis C virus (HCV) protease using structure-based computational modeling applying inverse design methods to implement the substrate envelope hypothesis. The protease structure was prepared for modeling and design studies using standard approaches. The other important residues that constituted the active substrate to be cleaved were also successfully modeled and substrate envelope is created. According to the synthesis requirement, the scaffold and building blocks are prepared for the design. 8. Effects of Macromolecular Crowding on Stem Cells(Krystyn Van Vliet (MIT/SMART-BioSyM), Michael Raghunath (NUS), Felicia C.Loe (SMART-BioSyM) and Adam Zeiger (MIT)Mesenchymal stromal or stem cells (MSCs) are a type of pluripotent precursor cell found in bone marrow, which can be coaxed in vitro to express certain characteristics of differentiated mesenchymal tissue lineages. Many therapeutic applications of MSCs require isolation from a given patient, expansion to useful cell numbers in culture, and reimplantation. Despite increasing clinical use, several fundamental questions of MSC in vitro studies and in vivo applications remain unanswered. Contributing to the difficulty in answering these questions are the distinct conditions of MSCs in vitro and in vivo. In vivo, the aqueous microenvironment of the cell is a crowded, protein rich, environment. In vitro, however, the extracellular culture media is decidedly dilute. Here, we utilize multiple methods to consider whether differential crowding can directly alter MSC structure and function. We have quantified changes in proliferation and migration, in structure and cortical cytoskeletal stiffness using immunohistochemistry, optical microscopy, and atomic force microscopy-enabled imaging and nanoindentation of living mesenchymal stem cells (MSCs) in basal media supplemented with macromolecular crowders. A preliminary analysis with immunohistochemistry has been carried out for fetal MSCs, to enable comparison with adult MSCs and pericytes.The results of the studies may help to create more physiologically relevant in vitro studies and biodevices, to ultimately help to answer fundamental questions of physiological and pathological behavior for MSCs. Selected publications
9. Macromolecular crowder amplifies adipogenic differentiation of mesenchymal stem cells through cell-matrix reciprocity(Krystyn Van Vliet (MIT/SMART-BioSyM), Michael Raghunath (NUS), Felicia C.Loe (SMART-BioSyM) and Adam Zeiger (MIT)The goal is to emulate the physiologically crowded cell environment by introducing inert macromolecules which in turn enhances extracellular matrix deposition and assembly and thus drives the differentiation of mesenchymal stem cells. We have used macromolecular crowding to amplify the adipogenic differentiation response of human bone marrow MSCs and showed that this amplification was brought about by the crowding-enhanced deposition and supramolecular assembly of ECM components along with enhanced subsequent lineage-specific remodeling of the ECM during differentiation. Selected publications
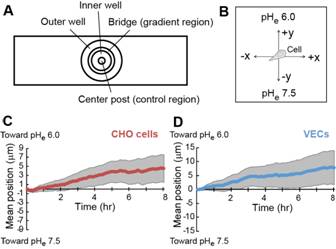
10. Directional cell migration in an extracellular pH gradientRanjani K. Paradise (MIT), Matthew J. Whitfield (MIT/SMART), Douglas A. Lauffenburger (MIT), and Krystyn J. Van Vliet (MIT/SMART) Extracellular pH (pH(e)) gradients are characteristic of tumor and wound environments. Cell migration in these environments is critical to tumor progression and wound healing. While it has been shown previously that cell migration can be modulated in conditions of spatially invariant acidic pH(e) due to acid-induced activation of cell surface integrin receptors, the effects of pH(e) gradients on cell migration remain unknown. Here, we investigate cell migration in an extracellular pH(e) gradient, using both model α(v)β(3) CHO-B2 cells and primary microvascular endothelial cells. For both cell types, we find that the mean cell position shifts toward the acidic end of the gradient over time, and that cells preferentially polarize toward the acidic end of the gradient during migration. We further demonstrate that cell membrane protrusion stability and actin-integrin adhesion complex formation are increased in acidic pH(e), which could contribute to the preferential polarization toward acidic pH(e) that we observed for cells in pH(e) gradients. These results provide the first demonstration of preferential cell migration toward acid in a pH(e) gradient, with intriguing implications for directed cell migration in the tumor and wound healing environments.
Selected publications
|
|
||||||||||||||||||||
|
|
|||||||||||||||||||||
BioSyM IRG
Publications Useful Links Global Enterprise for Micro Mechanics and Molecular Medicine (GEM4): www.gem4.org Mechanobiology Institute: http://mbi.nus.edu.sg/ Singapore-MIT Alliance for Research and Technology (SMART): http://smart.mit.edu/home.html
|
|||||||||||||||||||||